Hi All,
I’m trying to develop a better understanding of the amortized bayesian inference procedures and maybe compare them with vanilla bayesian modelling and/or VAE methods.
So i thought i’d try and do a parameter recovery exercise on a multivariate normal outcome. I have code working that i think is doing what it should do, but the estimation is super slow and for this “simple” example i feel i must have gotten something wrong in the set up. Could you advise if i’ve got the procedure right?
Generate Data
import pandas as pd
import numpy as np
import bayesflow as bf
# Standard deviations
stds = np.array([0.939, 1.017, 0.937, 0.562, 0.760, 0.524,
0.585, 0.609, 0.731, 0.711, 1.124, 1.001])
n = len(stds)
# Lower triangular correlation values as a flat list
corr_values = [
1.000,
.668, 1.000,
.635, .599, 1.000,
.263, .261, .164, 1.000,
.290, .315, .247, .486, 1.000,
.207, .245, .231, .251, .449, 1.000,
-.206, -.182, -.195, -.309, -.266, -.142, 1.000,
-.280, -.241, -.238, -.344, -.305, -.230, .753, 1.000,
-.258, -.244, -.185, -.255, -.255, -.215, .554, .587, 1.000,
.080, .096, .094, -.017, .151, .141, -.074, -.111, .016, 1.000,
.061, .028, -.035, -.058, -.051, -.003, -.040, -.040, -.018, .284, 1.000,
.113, .174, .059, .063, .138, .044, -.119, -.073, -.084, .563, .379, 1.000
]
# Fill correlation matrix
corr_matrix = np.zeros((n, n))
idx = 0
for i in range(n):
for j in range(i+1):
corr_matrix[i, j] = corr_values[idx]
corr_matrix[j, i] = corr_values[idx]
idx += 1
# Covariance matrix: Sigma = D * R * D
cov_matrix = np.outer(stds, stds) * corr_matrix
columns=["JW1","JW2","JW3", "UF1","UF2","FOR", "DA1","DA2","DA3", "EBA","ST","MI"]
corr_df = pd.DataFrame(corr_matrix, columns=columns)
cov_df = pd.DataFrame(cov_matrix, columns=columns)
cov_df
def make_sample(cov_matrix, size, columns):
sample_df = pd.DataFrame(np.random.multivariate_normal([0]*12, cov_matrix, size=size), columns=columns)
return sample_df
sample_df = make_sample(cov_matrix, 263, columns)
sample_df.head()
Which gives us a data set like:
What i’m interested in doing is estimating the covariance parameters which generated that sample. So i tried following the docs and set up my simulator and adapter etc so that i can do diagnostic tests and then ultimately calibrate the model against the observed sample data.
def random_corr(dim):
A = np.random.randn(dim, dim)
Q, _ = np.linalg.qr(A) # orthogonal matrix
R = Q @ Q.T # PD correlation-like
D = np.diag(1 / np.sqrt(np.diag(R)))
return D @ R @ D
def random_cov_lkj(dim):
R = random_corr(dim)
stds = np.exp(np.random.randn(dim)) # log-normal stds
D = np.diag(stds)
return D @ R @ D
def likelihood(mu, sigma):
N = sample_df.shape[0]
dim = sample_df.shape[1]
Sigma = sigma.reshape(dim, dim)
return {"y": np.random.multivariate_normal(mu, Sigma, size=N)}
def prior():
dim = sample_df_missing.shape[1]
mus = np.random.normal(0, 1, size=dim)
sigma = random_cov_lkj(dim)
return {"mu": mus, "sigma": sigma.ravel()}
simulator = bf.make_simulator([prior, likelihood])
sims=simulator.sample(1)
adapter = (
bf.Adapter()
.as_set(["y"])
.convert_dtype("float64", "float32")
.concatenate(["mu", "sigma"], into="inference_variables")
.concatenate(['y'], into='summary_variables')
)
summary_network = bf.networks.SetTransformer(summary_dim=10)
workflow = bf.BasicWorkflow(
simulator=simulator,
adapter=adapter,
summary_network=summary_network,
inference_network = bf.networks.CouplingFlow(),
inference_variables="y",
inference_conditions=["mu", "sigma"],
initial_learning_rate=1e-3,
standardize=None
)
training_data = workflow.simulate(1000)
validation_data = workflow.simulate(300)
## Runs but slowly
history = workflow.fit_offline(data=training_data, epochs=10, validation_data=validation_data);
bf.diagnostics.loss(history);
This code runs but does so quite slowly. Have i parameterised this poorly? I tried to recreate sampling of the covariance rather than say install pymc and sample from the LKJ prior… but i guess i could have done so poorly or inefficiently.
Ultimately i’m more interested in ensuring that the calibratio step works to estimate the posterior
post = workflow.sample(num_samples=200, conditions={'y': sample_df.values[None, :]})
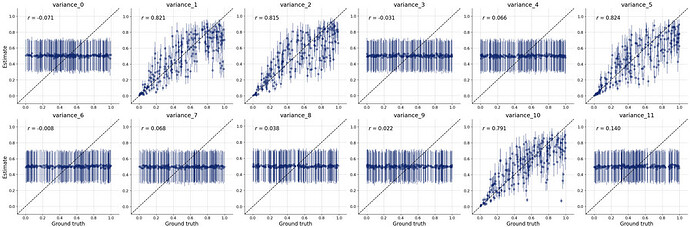
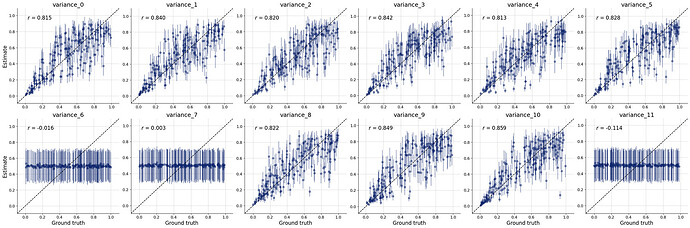
But i’m not sure if i’ve set up the whole workflow properly so i don’t really trust the posterior estimates. The diagnostic checks on the online process seem to suggest that it’s poorly specified too
Would you be able to advise if i’m on the right track? Or where i should go to ask for pointers on this use case? Thanks for your hard work and this cool looking package!